Autophagy
 Autophagy (literally “self-eating”) is the body’s way of cleansing cells by recycling old or damaged components and is a process that appears to have a strong relationship to preventing disease and aging. More technically, it is a highly regulated lysosome-dependent catabolic program used by the body to clear dysfunctional proteins, organelles and other structures that are typically first marked for the process by a protein called “ubiquitin.” Primarily intra-cellular but also extra-cellular components are engulfed in autophagocytic vacuoles and degraded to constituent molecules, such as sugars, fatty acids and amino acids, which can be then be used as nutrition to produce ATP and/or provide building blocks for the synthesis of essential proteins. There are different types of autophagy but for the most part they all serve this same purpose. Mitophagy is a similar process that removes damaged or dysfunctional mitochondria. There is also nucleophagy of material in the cell nucleus.
Autophagy (literally “self-eating”) is the body’s way of cleansing cells by recycling old or damaged components and is a process that appears to have a strong relationship to preventing disease and aging. More technically, it is a highly regulated lysosome-dependent catabolic program used by the body to clear dysfunctional proteins, organelles and other structures that are typically first marked for the process by a protein called “ubiquitin.” Primarily intra-cellular but also extra-cellular components are engulfed in autophagocytic vacuoles and degraded to constituent molecules, such as sugars, fatty acids and amino acids, which can be then be used as nutrition to produce ATP and/or provide building blocks for the synthesis of essential proteins. There are different types of autophagy but for the most part they all serve this same purpose. Mitophagy is a similar process that removes damaged or dysfunctional mitochondria. There is also nucleophagy of material in the cell nucleus.
A number of types of autophagy are currently described.
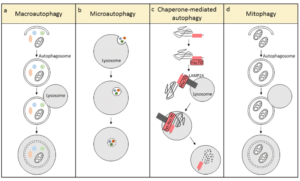
- Microautophagy: direct engulfment of cytoplasmic materials by invagination of lysosomal membrane.
- Macroautophagy: classic autophagasome mediated double membrane engulfment.
- Chaperone Mediated autophagy: extremely selective target protein mediated (Hsc70) recycling that is activated in response to cellular stress.
- Mitophagy: autophagy related to mitochondria and their components
- Also… ribophagy, lipophagy, xenophyagy, nucleophagy, clockophagy, lysophagy, etc.
Why is it important?
Autophagy is critical to cell function, regeneration and keeping the cell working properly. Inhibition of autophagy by knocking out any of a variety of autophagy related genes invariably accelerates aging and disease. Similarly, with aging, autophagy is downregulated (particularly in lymphocytes) leading to a forward feeding loop of accumulation of cellular debris and cellular dysfunction / dysregulation. In contrast, enhanced autophagy delays aging and prolongs lifespan in a range of model organisms including yeast, nematodes, fruit flies and mice. Lifespan studies in humans have not been conducted given the obvious challenges in conducting experiments in long-lived organisms, but it is generally assumed that regular induction of autophagy is a salubrious process, something we want to promote. So how to do so?
How to promote autophagy?
First the biochemistry–skip ahead to the punchline if you prefer. ULK1 is the enzyme that catalyzes production of phagosomes implicated in autophagy. AMPK and SIRT activate ULK1, MTOR deactivates it. If MTOR1 is inhibited by rapaymycin or starvation, autophagy is activated. TFEB is considered the primary transcription factor for the synthesis of both lysosome and autophagosomes. Again, AMPK boosts the activity of TFEB while MTOR inhibits it. Spermidine, which declines with age, is a required co-factor in the translation of TFEB, so its supplementation is theorized (and shown) to stimulate autophagy.
Caloric restriction activates autophagy by stimulating AMPK and sirtuin1 as well as inhibiting mTOR. When an autophagy related gene is knocked out or knocked down, the life-prolonging effect of rapamycin disappears, suggesting that autophagy plays an important role in the life-prolonging effect of rapamycin.
Simply not eating for a few hours and reducing nutrient levels in the blood starts the process of autophagy. This is the rationale for time restricted feeding. Starvation for longer periods such as two- or three-days initiates chaperone mediated autophagy, a targeted deep clean that modulates diverse functions such as glucose and lipid metabolism, DNA repair, cellular reprograming and the cellular response to stress.
Drugs to stimulate autophagy
- Berberine and metformin both boost AMPK, thus phosphorylating and activating ULK1–> autophagy.
- Rapamycin and spermidine inhibit MTOR, activating autophagy and spermidine, as already mentioned, both blocks motor and also is a necessary cofactor in the translation of TFEB.
- Resveratrol and NR (nicotinamide riboside) theoretically target SIRT1, though evidence of their bioavailability after oral administration is questionable.
- Ferulic acid may up-regulate SIRT1 expression and is derived from Angelica sinensis or “female ginseng” and used in traditional Chinese medicine. It is also widely available as an additive in cosmetic skin creams. Anthrocyanins in the diet are converted to ferric acid, providing a rationale for their health value.
- Urolithin-A, a phytochemical derived from pomegranate has also been shown to boost SIRT1, as well as improve mitochondrial biogenesis.
- Aerobic exercise has also been shown to stimulate autophagy.
- It turns out these are all the usual suspects in the realm of anti-aging drugs and supplements, and while many of them have diverse actions, it may be that their action on the autophagy pathways is primary to their purported value as anti-aging interventions. Yet the effect sizes of these interventions are not known, which is to say that we don’t really know to what extent autophagy will be triggered by taking any of these.
Can there be too much autophagy? Perhaps yes. While normal or even supernormal levels of autophagy promote cellular health, excessive or uncontrolled autophagy can initiate a type of cell death known as “autosis” (https://www.nature.com/articles/cdd2014143). While autophagy is believed to be helpful, cell death through autosis is inarguably harmful and to be avoided. The clinical significance of autosis in the context of therapeutic autophagy is not clear. I reached out to Beth Levine, the woman who did much of the original research on autosis but her email bounced back and it turns out she has tragically died of breast cancer. In any case, in this setting of incomplete understanding, the best course is one of prudence and moderation. In the words of Hippocrates, first do no harm. Practically speaking, my policy is to cycle agents, to avoid excessive dosing and to carefully follow serologic markers during treatment.
Relationship between autophagy and weight?
There is an emerging understanding of a relationship between autophagy in the medial-basal-hypothalamus (MBH) and obesity mediated by the FK506 (tacrolimus) binding protein FKBP51, which appears to be a major upstream regulator of autophagy. Could autophagy and more broadly, the intracellular environment in hypothalamic neurons be the functional memory that induces phenomena such as the physiologic set point? Could agents that augment autophagy such as rapamycin reset the physiologic set point in humans, as it has in mouse models? These are intriguing questions.

 How do we measure aging? Certainly, there is the passage of time and how many birthdays someone has had, but we know that aging proceeds more quickly for some people than it does for others. The speed of aging depends on your genes but also various factors such as, for instance, whether you smoke, your diet and level of exercise, your stress levels, exposure to ionizing radiation, and many other variables. A 50 year old executive will likely be biologically younger than a 50 year old homeless alcoholic who has had a hard life, to take an example. Is there some other marker that indicates a biological age that is distinct from chronological age? This question is critically important if we are to assess the effectiveness of anti-aging interventions and has been a matter of much debate. Recently as our understanding of aging has evolved, scientists have focused on methylation patterns in the DNA as a likely proxy for aging.
How do we measure aging? Certainly, there is the passage of time and how many birthdays someone has had, but we know that aging proceeds more quickly for some people than it does for others. The speed of aging depends on your genes but also various factors such as, for instance, whether you smoke, your diet and level of exercise, your stress levels, exposure to ionizing radiation, and many other variables. A 50 year old executive will likely be biologically younger than a 50 year old homeless alcoholic who has had a hard life, to take an example. Is there some other marker that indicates a biological age that is distinct from chronological age? This question is critically important if we are to assess the effectiveness of anti-aging interventions and has been a matter of much debate. Recently as our understanding of aging has evolved, scientists have focused on methylation patterns in the DNA as a likely proxy for aging.